Plant Functional Genomics
Research Groups
Group of Genetic Resources and FunctionsStaff
Research Topics
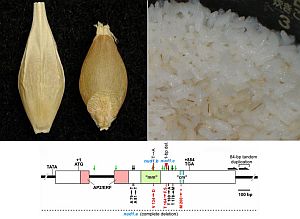
Barley, the fourth most important cereal crop in the world, typically has seeds with adhered hulls. This hulled seed trait is suitable for breweries. Some naked seed mutants with easily separable hulls were generated through spontaneous mutation. They are suitable for food usage. We have revealed that barley awns and spike hulls are photosynthetically active. Our current research specifically examines molecular identification and characterization of beneficial genes controlling (1) seed morphology and quality, including water soluble healthy dietary fiber, and (2) photosynthetic contribution of awns and hulls by using mutants. We seek application of our basic findings to practical breeding.
Molecular dissection of beneficial genes in barley
Diversity in plant and flower morphology of barley
Exploitation of Non-foliar photosynthesis and viral disease resistance for sustainable barley production
Publication List
- (1) Taketa, S., Hattori, M., Takami, T., Himi, E, Sakamoto, W. Mutations in a Golden2-like gene cause reduced seed weight in barley albino lemma 1 mutants. Plant and Cell Physiology 62: 447-457 (2021) (2021)
- (2) Hibara, K-I., Miya, M., Benvenuto, S.A., Hibara-Matsuo, N., Mimura, M., Yoshikawa, T., Suzuki, M., Kusaba, M., Taketa, S., Itoh, J. Regulation of the plastochron by three MANY-NODED DWARF genes in barley. PLOS Genetics 17(5): e1009292 (2021)
- (3) T., Yoshikawa, H., Hisano, K-I. Hibara , J. Nie , Y. Tanaka , J-I. Itoh , S. Taketa. A bifurcated palea mutant infers functional differentiation of WOX3 genes in flower and leaf morphogenesis of barley. AoB Plants (2022, in press)
- (4) Ube, N., Katsuyama, Y., Kariya, K., Tebayashi, S., Sue, M., Tohnooka, T., Ueno, K., Taketa, S., Ishihara, A. Identification of methoxylchalcones produced in response to CuCl2 treatment and pathogen infection in barley. Phytochemistry 184: 112650 (2021).
- (5) Fukunaga, K., Nur, M.Z., Inoue, T., Taketa, S., Ichitani, K. Phylogenetic analysis of the Si7PPO gene in foxtail millet, Setaria italica, provides further evidence for multiple origins of the negative phenol color reaction phenotype. Genes and Genetic Systems 95: 191-199 (2020).
- (6) Milner, S.G., Jost, M., Taketa, S., Mazón, E.R., Himmelbach, A., Oppermann, M., Weise, S., Knüpffer, H., Basterrechea, M., König, P., Schüler, D., Sharma, R., Pasam, RK., Rutten, T., Guo, G., Xu, D., Zhang, J., Herren, G., Müller, T., Krattinger, S.G., Keller, B., . Jiang, Y., Gonzalez, M.Y., Zhao, Y., Habekuß, A., Färber, S., Ordon, F., Lange, M., Börner, A., Graner, A., Reif, J.C., Scholz, U., Mascher, M. and Stein, N. Genebank genomics highlights the diversity of a global barley collection. Nature Genetics 51: 319-326 (2019).
- (7) Jost, M.,* Taketa, S.,* (*co-first authors), Mascher, M., Himmelbach, A., Yuo, T., Shahinnia, F., Rutten, T., Druka,A., Schmutzer, T., Steuernagel, B., Beier, S., Taudien, S., Scholz, U., Morgante, M., Waugh, R. and Stein, N. A homolog of Blade-On-Petiole 1 and 2 (BOP1/2) controls internode length and homeotic changes of the barley inflorescence. Plant Physiology 171: 1113-1127 (2016).
- (8) Ube, N., Harada, D., Katsuyama, Y., Osaki, K., Tonooka, T., Ueno, K., Taketa, S. and Ishihara, A. Identification of phenylamide phytoalexins and characterization of inducible phenylamide metabolism in wheat. Phytochemistry 167:112098 (2019).